Health Systems
Deliver explainable medication actions directly in clinical workflow.
Deliver explainable medication actions directly in clinical workflow.
Enable labs with DGI signals inside a unified medication decision layer.
Accelerate translational and trial programs with unified medication insights.
Evidence-informed coverage decisions tied to measurable real-world outcomes.
Plain-language medication guidance for safer, better-informed care conversations.
Unified medication-response reasoning with explainable, evidence-linked output.
EHR-native delivery through FHIR/HL7/CDS integration pathways.
Governance, auditability, versioning, and safe rollout controls.
A clear path from pilot to enterprise rollout inside the health system.
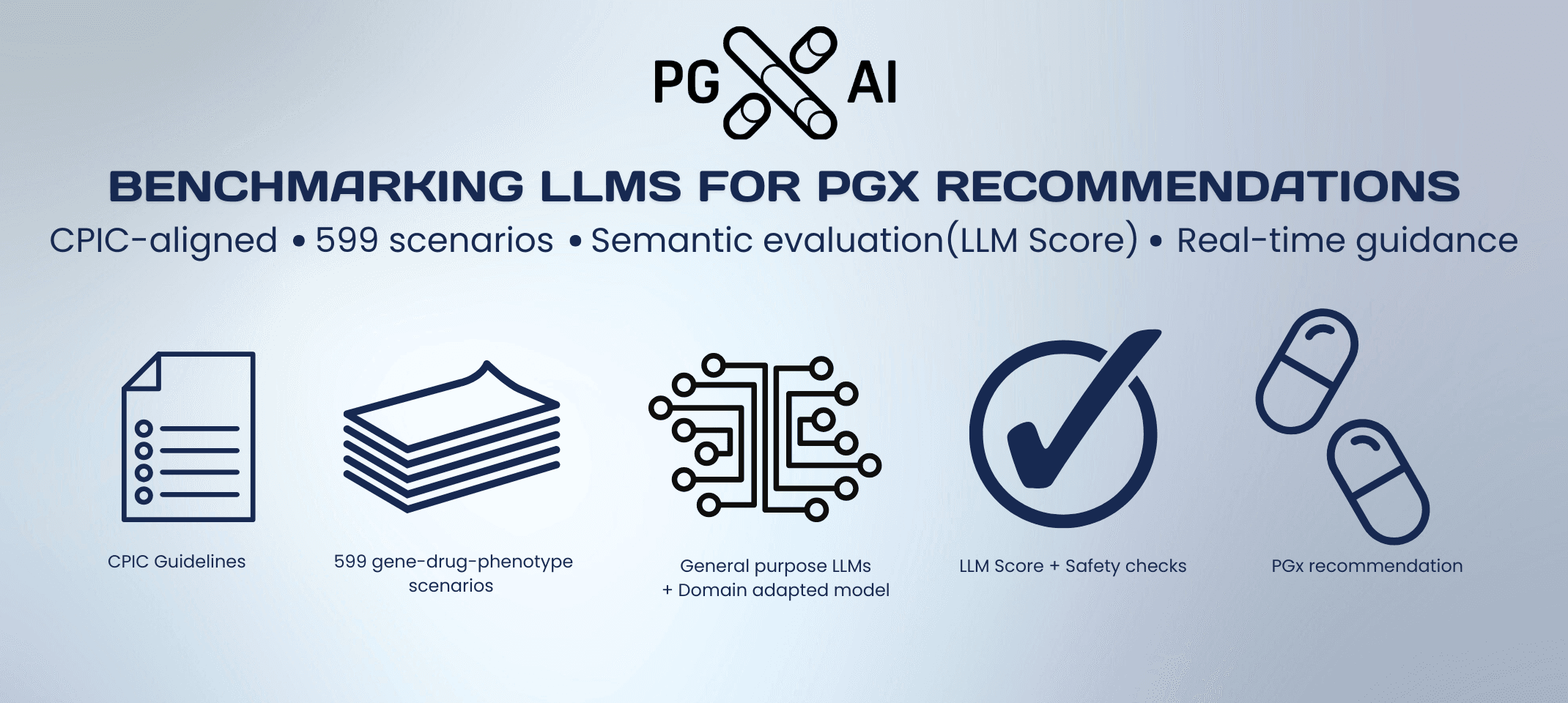
PGx testing can markedly improve treatment safety and efficacy, yet adoption in health systems remains slow. Our latest peer-reviewed study-now published in The Pharmacogenomics Journal (Nature Portfolio)-benchmarks how well large language models (LLMs) reproduce guideline-based pharmacogenomic (PGx) recommendations and outlines a practical path from static, one-time advice to dynamic guidance updated in real time.
Nature
Key findings
General-purpose models often produced incomplete or unsafe outputs when asked for guideline-conformant PGx recommendations.
Domain adaptation (including targeted fine-tuning and structured prompting) substantially improves accuracy and reduces latency, making PGx-specific LLMs more suitable for clinical decision support.
Why this matters
LLMs purpose-built for PGx can help close the gap between genetic results and bedside actionability-supporting safer, faster, and more consistent medication decisions across diverse patient populations.
Looking ahead
We have already released a closed model, Deneb, capable of generating recommendations in real time. Comparative testing against the latest versions (o3, o3-pro, grok-4, claude-4, etc.) will be shared soon.