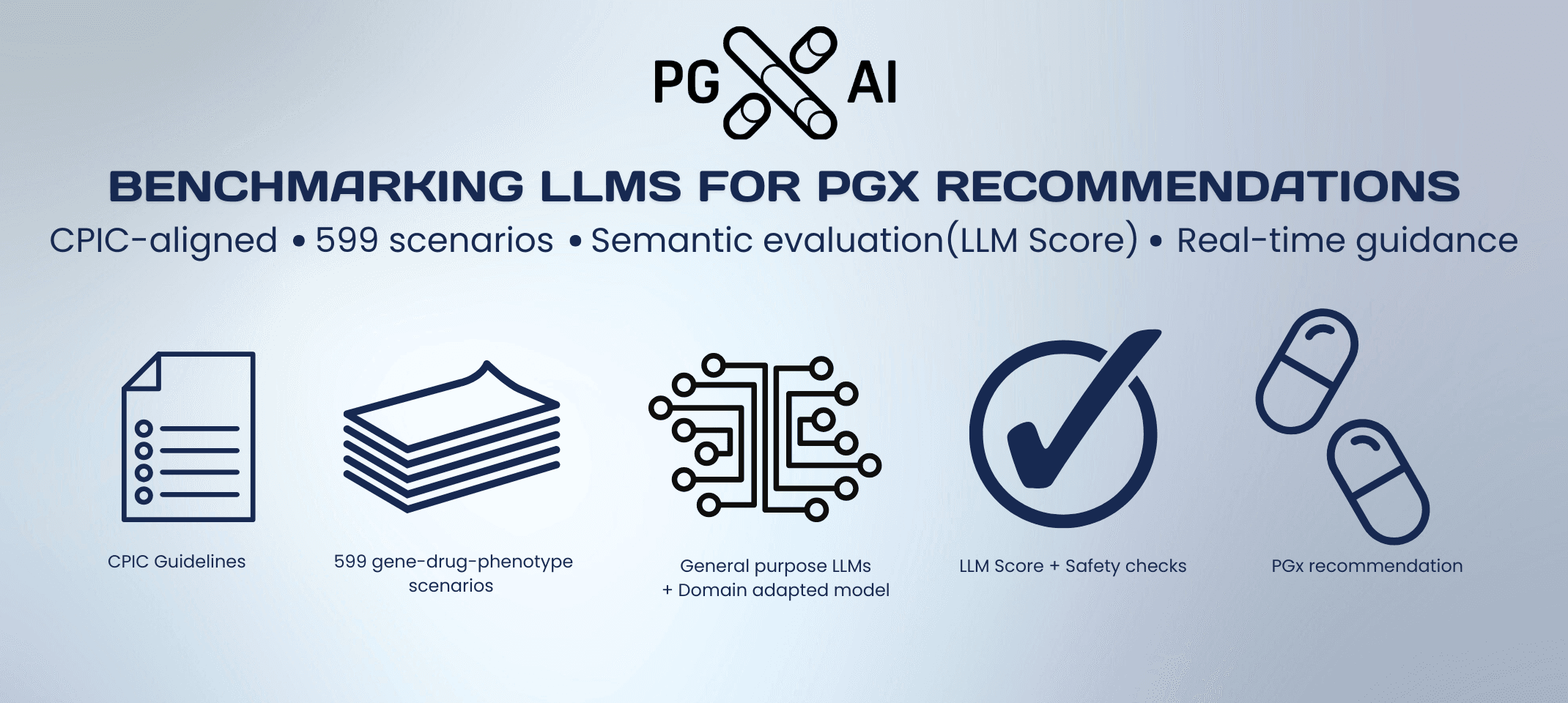
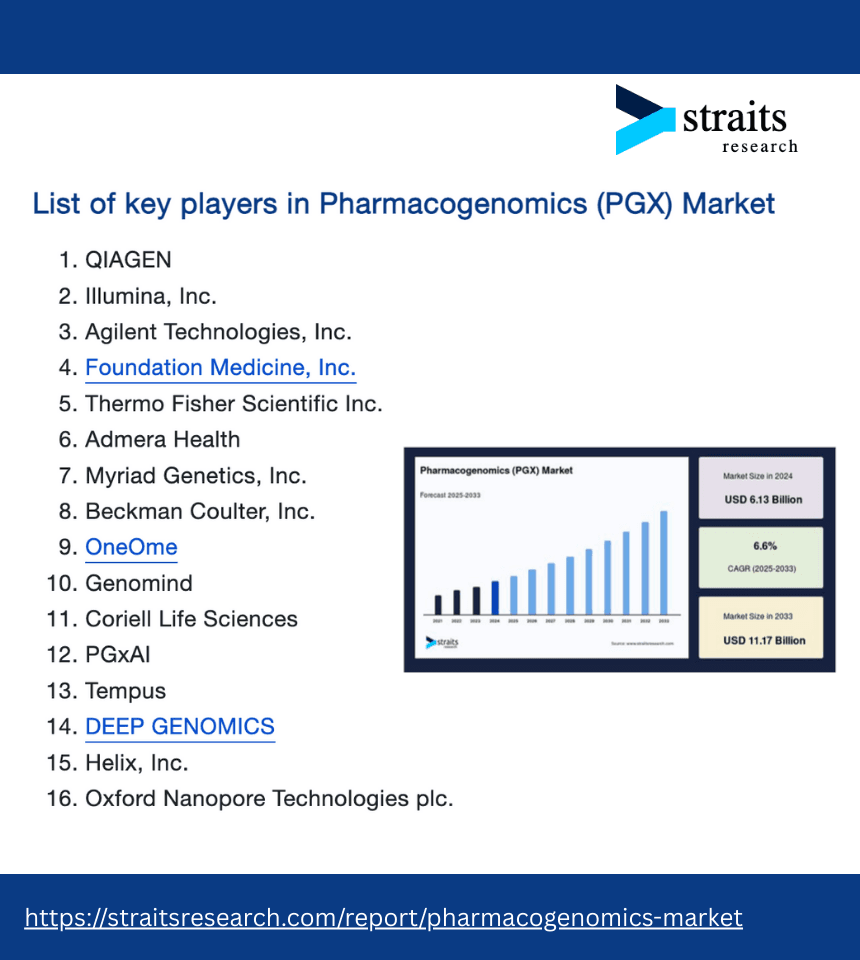
PROVIDERS
LABS
CROs
PAYORS
PATIENTS
ABOUT US
Therapeutic Areas
LABS
Overview LIMS Integration Implementation & Workflow Expand Your Test Offerings Case Studies & Testimonials Lab FAQ Contact / Request a Demo Genomic Reports
CROs
Overview Trial Optimization & Patient Stratification Technology & Workflow Integration Multi-Phase Support Implementation Steps CROs FAQ PAYORS
Optimizing Coverage with Precision Medicine Cost Savings Through Precision Evidence-Based Coverage Seamless Integration with Payor Systems Value-Based Care Alignment PATIENTS
Overview How It Works Coverage & Reimbursement Conditions & Medications Resources & Education Privacy & Data Security Request a Demo ABOUT US
Overview Our Story Our Team Our Technology Culture & Careers Achievements & Growth Partnerships & Market Presence Press Releases & News Documents & Presentations Valuation & Market Potential FAQ Email Alerts & Contact Menu
PROVIDERS
LABS
CROs
PAYORS
PATIENTS